Introduction
While global pandemics have been a common occurrence for much of human history, their damage was limited to the small geographical regions where the disease originated. Now, with the rapid globalization of the world, a disease originating in a remote province in China or Mexico can spread across the globe in a matter of a few months, causing thousands of lives (Flahault 2010, 320). In an effort to contain a new disease in an early phase before it becomes a global pandemic, health agencies use advanced surveillance techniques to monitor developments in infections around the world.
The methodology employed by these systems is focused on being able to detect when and where a serious case might have been reported, by scraping news stories from various sources on the internet and looking for symptoms that might indicate an outbreak (Government of Canada 2004). This allows the systems to be able to alert the responsible authorities in the first few weeks of an outbreak. While this technique is effective in its goal of detecting diseases before they become pandemics, it misses an opportunity to achieve a larger objective – predicting areas where an outbreak is imminent and containing it even earlier.
“HealthMap identified a new pattern of respiratory illness in Mexico in 2009 well before public-health officials realized a new influenza pandemic was emerging. Yet, researchers didn’t recognize the seriousness of the threat it posed.” – Dr. Brownstein, Creator of HealthMap (Weeks 2011)
A quick study of the pandemics over the last decade indicate that while global disease monitoring systems did alert the authorities within the first few weeks of an outbreak, it was already too late to contain it (Weeks 2011). Therefore, a system that can identify where the next disease outbreak is most likely to occur, will be more effective in obtaining these objectives. The methodology required to make such predictions is not new, but already used in several other applications, such as predicting movie sales based on sentiment analysis of social media feeds and predicting stock prices, based on how a new product or service is received by the consumers. In a similar technique, a semantic analysis of the content from the web, combined with a knowledge base of data that is trained to look for clues of disease outbreaks based on historical patterns can be used to predict where a disease outbreak is most likely.
Current Disease Surveillance Systems
Global and national health agencies have developed systems that they use to scan the content on the internet for threats they believe might develop into global pandemics. These systems rely on complex software to perform this scan because of the amount of content that needs to be monitored. As the system scans the internet, it produces results that are of interest to the analysts, that study it further to determine if it is a genuine threat (Government of Canada 2004).
“It’s still a lot of information, a lot of noise. In hindsight, you can say this was the first indication, but … when you look at all these reports coming in from all over the world on a daily basis, it is hard or impossible to tell which of these reports constitutes a real threat.”
– Dr. Gunther Eysenbach, University of Toronto’s Centre for Global eHealth Innovation (Blackwell 2009)
Global Public Health Intelligence Network
Global Public Health Intelligence Network (GPHIN) was developed by the Public Health Agency of Canada and is the world’s premier disease surveillance system. Its objective is to scour the web using a text mining engine to look for symptoms of diseases or reports of contaminated food or water supplies and, if necessary, generate an alert to an analyst. The GPHIN system uses a text mining engine developed by Nstein that is also capable of translating news articles between 7 different languages, to maximize its sources of data (Government of Canada 2004). This system has been successful in detecting more than half of the 578 outbreaks identified by the World Health Organization between 1998 and 2001. It is also responsible for being the first disease surveillance system to detect news stories of an outbreak of SARS and Avian Flu, in the very early stages, from the Chinese and the Mexican media respectively (Blackwell 2009).
HealthMap
The HealthMap project, developed by researchers at the Children’s Hospital Boston, has a similar objective. Their software system scours the web, news websites, government and non-government organizational websites and social media feeds to collect data that it analyzes to produce a visualization of the current state of the diseases around the world (Freifeld 2008, 151). The system is capable of mining content from 9 different languages. It filters the content based on keywords of symptoms of diseases common in humans, animals and plants, then further extracts the time and location information from the source. Finally, it uses this data to generate a map of the world, highlighting those regions that have the most cases reported of a specific disease (Freifeld 2008, 152).
“Early detection of disease activity, when followed by a rapid response, can reduce the impact of both seasonal and pandemic influenza. One way to improve early detection is to monitor health-seeking behavior in the form of online web search queries, which are submitted by millions of users around the world each day.” (Ginsberg 2008, 1)
Google Flu Trends
Google’s Flu Trends is another prominent disease detection system. It has a slightly different objective from GPHIN and HealthMap – it is limited to identifying areas of high flu activity. Their methodology towards generating the map of their results is also very different from GPHIN and HealthMap. Rather than using any complex semantic analysis software, Google Flu Trends focuses on studying patterns in Google web searches to determine what areas might be experiencing a spike in flu related cases. This information is then transmitted to the local health agencies and hospitals which can prepare for the spike in cases and flu vaccines. Google Flu Trends currently works in 15 countries and only relies on data from Google search queries (Ginsberg 2008, 1).
Historical Approach
“Outside experts say programs like it [GPHIN] and Harvard Medical School’s HealthMap are useful, but only when combined with other sources of information that, pieced together, paint a bigger picture.” (Blackwell 2009)
The traditional approaches to the problem of containing global pandemics has been to detect them as early as possible through the means of inter-agency co-operation at national and international levels. This meant that the agencies would share data between each other in an effort to develop an understanding of the current state of the diseases around the world.
This approach was refined with the advent of the internet, as large amounts of data was available for study and analysis. Organizations made use of text mining techniques and semantic analysis to data mine the internet for articles relevant to their research, and analyze them further (Weeks 2011). The current methodology common to most systems can be described in the process below (Freifeld 2008, 152).
-
Scraping – Mine news websites, government and NGO websites for textual information containing keywords of symptoms of diseases or relevant phrases from a predetermined dictionary database.
-
Data extraction – Extract the useful information from the text, such as the title, author, source, date, body and the URL.
-
Semantic Analysis – Extract information from the content about its location, time, and disease related information, such as symptoms and description.
-
Generate Result – The data produced is transferred to analysts that examine it further to assess the risk posed.
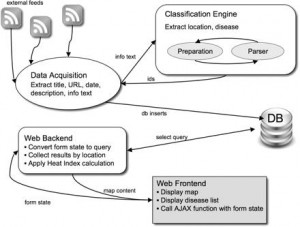
Figure. 1 HealthMap System Architecture
Figure 1 illustrates the architecture used by HealthMap to generate the map of areas suffering from disease outbreaks. Most systems that use this methodology are able to analyze sources from multiple languages by using language translation tools and dictionaries in the second step (Freifeld 2008, 153). Such an approach proves to be quiet effective in filtering the content and producing relevant results. For example, using this methodology, GPHIN was able to detect a news story out of Guangdong province of China, within weeks of the first SARS case, as a outbreak of a “respiratory” disease (Blackwell 2009). It was also able to detect a news story out of Mexico in 2009 that reported deaths caused by a “strange epidemic outbreak” of a flu like illness, weeks before it was known as H1N1 flu to the world (Ibid).
Pitfalls of the Historical Approach
The objective of developing these advanced software systems is to be able to contain the spread of a new disease in as early a phase as possible, before it becomes a global pandemic (Government of Canada 2004). However, the historical approach to solving this problem has been focused on detecting the millions of cases of diseases around the world, and then analyzing them further by time, location, severity and symptoms to determine if they are a global threat. This approach dilutes the original objective of preventing disease outbreaks, and re-channels it towards detecting disease outbreaks, and then alerting the authorities. This approach clearly does not address the underlying problem, because even though it is able to make use of technology and the data available to us to detect a disease outbreaks weeks before it is a pandemic, it does not go far enough in taking advantage of the technologies and data available to us.
The scope of the data mined by historical approaches is limited to a few languages and sources such as large public news outlets. Due to this, the system inherits a bias in its data towards certain regions of the world.
Data Sources
One of the biggest shortcomings of the historical approach is its limited scope of data sources. The system can inherit a bias into its content based on the data sources it monitors, and the languages it supports. If the data source includes RSS feeds of prominent news outlets, social media feeds and search queries, as most of the current systems do, the system is ignoring a large section of the content that could potentially contain very significant information. A lot of the reports that contain information that can help determine areas of concern would come from reports published by non-governmental agencies, aid agencies, other international monitoring agencies, talks at a conference and even blogs. These sources might not include reports of actual cases of a disease outbreak, but a study of their content can certainly reveal signs that an outbreak might be imminent. For example, Doctors Without Borders might publish a report detailing deteriorating water standards in a specific region of the world, which might get overlooked by current text mining engines because it does not contain certain keywords or phrases, however, the information gathered from it might lead an intelligent system to flag such an event for a possible area of a disease outbreak.
The text mining engines used by current surveillance systems are limited in their functionality to getting the time and location information of an event, rather than learning new facts from the context of the article.
Semantic Analysis
The methodology of the historical approaches uses the advances in natural language processing in a very limited way. While they use a data mining engine to learn time and location information from a data source where it found matches for certain keywords or phrases, it fails to develop a knowledge base of facts that it can then use to perform a more effective analysis of a situation. It also fails to perform a thorough semantic analysis on the original content to scan for information that could be relevant to a disease outbreak, by just performing a keyword analysis (Freifeld 2008, 157).
Threat Analysis
The methodology for threat analysis differs from system to system, however none of them is advanced enough to analyze the data by comparing it to a knowledge base of facts, and determine how severe or unusual a threat might be (Weeks 2008). For example, while GPHIN was able to detect the story about the outbreak of SARS in 2002 from a Chinese media source, it failed to perform an automated threat analysis of the story, and was not successful in alerting the global health agencies in time to contain the outbreak within the province of Guangdong (Ibid).
Predict, not detect
“The next noteworthy outbreak may as easily come from a major urban center in North America as a rural village in Africa.” (Freifeld 2008, 152)
A smarter and more effective approach to preventing an outbreak of a global pandemic is to be able to predict which region of the world is the most likely breeding ground for one. In such a scenario, prevention of the disease could precede cases of the disease outbreak being reported. While it is true that certain parts of the world will always be more prone to an outbreak because of the lack of public health infrastructure or the sanitary conditions that persist, but a new strain of a disease that has the potential to spread globally as a pandemic can come from any part of the world. “The next noteworthy outbreak may as easily come from a major urban center in North America as a rural village in Africa. ” (Freifeld 2008, 152).
Determining Factors
Parts of the world that are at high risk of an epidemic could be determined by factors such as animal farming practices in a local community (Cyranoski 2008), or food growing practices, sanitary conditions, quality of water and food sources (Piarroux 2011, 1162), civil unrest and even a humanitarian crisis. A system that is able to monitor the internet for these factors, and build a knowledge base of facts that it uses to analyze the reports will be far more effective in reaching the objectives, than a system that relies on detecting an outbreak from news stories after it has occurred. This has been true for SARS and Avian flu, where the conditions for the birth of such a disease could have been predicted had a system been in place to monitor for such factors.
“The concentration of humans or animals in proximity enhances potential transmission of microorganisms among members of the group. It also creates greater potential for infecting surrounding life forms, even those of different species. The conditions created also may be a breeding ground for new, more infectious, or more resistant microorganisms. ” (Gilchrist 2006, 313)
Methodology for a Predicting System
Developing a knowledge base of facts and a Bayesian network to determine what factors could indicate an outbreak can vastly improve the effectiveness of disease surveillance systems.
The goal of predicting disease outbreaks rather than detecting them is not beyond the reach of current technological capabilities. Software systems that are able to filter the data for facts and use it to perform a sophisticated analysis already exist, and are commonly used in other applications such as predicting civil uprisings (Leetaru 2011), consumer behavior (Harrison 2012) and even movie ticket sales (Yu 2012, 720).
Knowledge Base
To be able to apply a similar approach in the case of disease prevention requires a modification to the current methodology and the software architecture. The new software architecture will be required to have a separate database of facts, that can be manually fed into the system, or that can be “learned” from a data source, called a Knowledge base. This component of the design will be responsible for making informative decisions based on the data it gathers and the facts it already knows. It will be accessible by the team of medical researchers and technical staff that maintain the system, so that they can update the knowledge base accordingly.
The knowledge base will consist of facts that it needs to make decisions, such as:
- Factors that can onset a disease outbreak
- Patterns of a historical disease outbreak
- Bayesian network of symptoms, their classification and relations to diseases
- Bayesian network of diseases and their relation to age, gender and elasticities
- Economic data of a country – Gini index, accessibility to remote areas of the country
- Data on the public health institutions – number of hospitals, number of doctors per people
- Data on political stability
The knowledge base is the most significant modification to the existing methodology. It will evolve as it learns more data and improve its prediction algorithm over time. It will act as a brain of the new system, that processes the information to be able to perform a threat analysis of a disease, a symptom or any event. For example, when this system learns of an earthquake in Haiti of a large magnitude, it will be able to process the geographical, economical and demo-graphical information through its knowledge base and predict that certain types of diseases that are common in conditions of deteriorating sanitary conditions are likely, such as Cholera, as was the case after the 2008 Earthquake (Piarroux 2011, 1162).
Data Source
One of the most important shortcomings of the current methodologies that needs to be addressed is the small scope of their data sources. The effectiveness of the new system is directly proportional to the breadth of data fed into it, because as it scours the data, it looks for facts and stores them in its knowledge base. Therefore, the more the system scours, the more it learns and the better its prediction algorithm. While increasing the data sources and the format of sources the system accepts to include blogs, non-governmental websites, hospital records, conference discussions, journals and social media feeds is not a challenge, it is quiet important to assure the quality of content reported from these sources. Therefore, the scanning engine will determine the source of the article, and look up its references and overtime develop a database of good quality sources that it will use to filter out its data gathering.
Semantic Analysis
The semantic analysis performed under this methodology is to look for more than just keywords of symptoms or phrases common in disease descriptions. It will also include a scan for factors that indicate a likely disease outbreak, such as news stories of bad animal or food farming practices or suspicious deaths. It will also scan for news stories of a humanitarian crisis in a region of the world which is not equipped to deal with it. It will scan the content and look for articles that might indicate factors that have been flagged in the knowledge base. The semantic analysis will also conduct a temporal and contextual analysis on the article to learn information about the time, location and the context that the event has occurred in. It will use facts from its knowledge base to improve the analysis. Most importantly, it is at this step that the semantic analysis engine will use a pattern dictionary approach to translate content from as many languages as it has capability in its language module for. It will look for words or phrases and using a dictionary translator, look for those keywords and phrases in the source from another language as well. Overtime, it will learn from this pattern dictionary approach and improve its language module of the semantic analysis engine as well.
Threat Analysis
This is one of the most significant steps of the methodology for a system that is able to make predictions. It is at this step that the system analyzes each data it has collected, and uses its contextual, language, location and time information to co-relate it with the information it has about it in the knowledge base, and produce a severity rating. The severity rating of each data is based on the analysis of its content, such as the event that has happened, and combining it with the facts it has in its knowledge base that allows the system to make intelligent predictions. The system will make use of the data it gathered from the semantic analysis and see if the facts in the knowledge base, such as the Bayesian network or other factors together indicate if this event poses a serious threat. If it does, it will assign it a rating based on the quality of the source, the historical pattern and the socio-economic indicators of the country.
Generate Results
An analysis of each event and the threat it poses requires the power of a large knowledge base working behind the scenes to be able to make predictions.
The final step of the methodology is to produce a visualization of the data so the analysts and the medical authorities can make use of it. HealthMap has a good front-end visualization of their results, that is shown in the figure below.
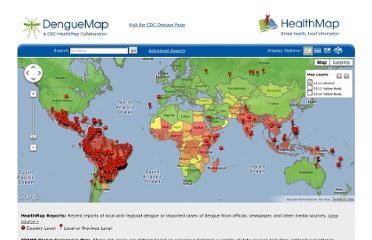
Figure. 2 Visualization of HealthMap’s Data
Conclusion
Considering the goals of a global disease surveillance system and the systems examined in this paper, several important conclusions can be reached. The current systems do not employ a methodology that can provide alerts with adequate time to contain the global spread of the disease (Weeks 2011). Their methodology involves monitoring a limited sources on the internet for keywords or phrases that could indicate a risk of a potential disease outbreak. However, a thorough semantic analysis of all content, that can look for factors to indicate where a disease outbreak might occur could make the system far more effective. This analysis combined with a knowledge base consisting of facts that would help the system process the data it gathers and rate it based on the threat it poses, is an indispensable component of any modern disease surveillance system. An advanced monitoring system that can use the knowledge base to make predictions can alert health authorities of a potential disease outbreak weeks before it occurs and thus save thousands of lives and billions of dollars.
Works Cited
Abbasi, Ahmed, Hsinchun Chen, and Arab Salem. 2008. ‘Sentiment Analysis in Multiple Languages: Feature Selection for Opinion Classification in Web Forums’. ACM Transactions on Information Systems. http://lcweb.senecac.on.ca:2134/10.1145/1370000/1361685/a12-abbasi.pdf? ip=142.204.1.85&acc=ACTIVE%20SERVICE&CFID=95479844&CFTOKEN=
34864361&__acm__=1333510662_0da55de99a 1cb0f3aaa2f528d6e2dd7f.
Blackwell, Tom. 2009. ‘Health Officials Aim to Hone Disease Surveillance’, May 3. http://www.nationalpost.com/news/canada/story.html?id=1562873.
Cyranoski, David. 2005. ‘Bird Flu Spreads Among Java’s Pigs’. Nature 435 (7041) (May 25): 390–391. doi:10.1038/435390a.
Flahault, Antoine and Patrick Zylberman. 2010. ‘Influenza Pandemics: Past, Present and Future Challenges’. BMC Infectious Diseases 10 (1): 162. doi:10.1186/1471-2334-10-162.
Freifeld, Clark C, Kenneth D Mandl, Ben Y Reis, and John S Brownstein. 2008. ‘HealthMap: Global Infectious Disease Monitoring Through Automated Classification and Visualization of Internet Media Reports’. Journal of the American Medical Informatics Association 15 (2) (January 3): 150–157. doi:10.1197/jamia.M2544.
Gilchrist, Mary J., Christina Greko, David B. Wallinga, George W. Beran, David G. Riley, and Peter S. Thorne. 2006. ‘The Potential Role of Concentrated Animal Feeding Operations in Infectious Disease Epidemics and Antibiotic Resistance’. Environmental Health Perspectives 115 (2) (November 14): 313–316. doi:10.1289/ehp.8837.
Ginsberg, Jeremy, Matthew H. Mohebbi, Rajan S. Patel, Lynnette Brammer, Mark S. Smolinski, and Larry Brilliant. 2008. ‘Detecting Influenza Epidemics Using Search Engine Query Data’. Nature 457 (7232) (November 19): 1012–1014. doi:10.1038/nature07634.
Government of Canada, Public Health Agency of Canada. 2004. ‘Global Public Health Intelligence Network (GPHIN) – Information – Public Health Agency of Canada’. http://www.phac-aspc.gc.ca/media/nr-rp/2004/2004_gphin-rmispbk-eng.php.
Harrison, Guy. 2012. ‘Sentiment Analysis Could Revolutionize Market Research’. Database Trends & Applications.
HealthMap System Architecture. Image.
2007. http://jamia.bmj.com/content/15/2/150.full#xref-ref-27-1 (accessed March 29, 2012).
Heffernan, Richard Farzad Mostashari, Debjani Das, Adam Karpati, Martin Kulldorff† and Don Weiss. ‘Syndromic Surveillance in Public Health Practice, New York City’. http://wwwnc.cdc.gov/eid/article/10/5/03-0646_article.htm.
Leetaru, Kalev H. 2011. ‘Culturomics 2.0: Forecasting Large–scale Human Behavior Using Global News Media Tone in Time and Space’. First Monday 16 (9) (August 17). http://www.uic.edu/htbin/cgiwrap/bin/ojs/index.php/fm/article/view/3663/3040.
Neill, Daniel B. 2012. ‘New Directions in Artificial Intelligence for Public Health Surveillance’. IEEE Intelligent Systems 27 (1) (January): 56–59. doi:10.1109/MIS.2012.18.
Piarroux, Renaud. 2011. ‘Understanding the Cholera Epidemic, Haiti’. Emerging Infectious Diseases 17 (7) (July): 1161–1168. doi:10.3201/eid1707.110059.
Recorded Future. 2011. ‘Big Data for the Future: Unlocking the Predictive Power of the Web’. Education October 18. http://www.slideshare.net/RecordedFuture/big-data-for-the-future- unlocking-the-predictive-power-of-the-web.
Subrahmanian, V.S., and Diego Reforgiato. 2008. ‘AVA: Adjective-Verb-Adverb Combinations for Sentiment Analysis’. IEEE Intelligent Systems 23 (4) (July): 43–50. doi:10.1109/MIS.2008.57.
Visualization of HealthMap’s Data. Image.
2012. http://www.healthmap.org (accessed April 03, 2012).
Weeks, Carly. 2011. ‘Social Media Could Help Detect Pandemics, MD Says’. The Globe and Mail. http://www.theglobeandmail.com/life/health/new-health/health-news/social- media-could-help-detect-pandemics-md-says/article2077719/.
Yu, Xiaohui1, Yang2 Liu, Xiangji3 Huang, and Aijun3 An. 2012. ‘Mining Online Reviews for Predicting Sales Performance: A Case Study in the Movie Domain’. IEEE Transactions on Knowledge & Data Engineering.
Zheng, Wanhong, Evangelos Milios, and Carolyn Watters. 2002. ‘Filtering for Medical News Items Using a Machine Learning Approach.’ Proceedings of the AMIA Symposium: 949– 953.